ITS_LIVE Global Glacier Velocity Exploration and Analysis ¶

Luis Lopez1, Alex Gardner2, Mark Fahnestock3, Ted Scambos4, Maria Liukis2, Chad Greene2, Yang Lei5, Joe Kennedy3, Bruce Wallin1
1.National Snow and Ice Data Center
2.NASA Jet Propulsion Laboratory
3.University of Alaska Fairbanks
4.University of Colorado Earth Science and Observation Center
5.California Institute of Technology
1.1 Authors ¶
Author1 = {“name”: “Luis Lopez”, “affiliation”: “National Snow and Ice Data Center”, “email”: “luis.lopez@nsidc.org”, “orcid”: “”}
Author2 = {“name”: “Alex Gardner”, “affiliation”: “NASA Jet Propulsion Laboratory”, “email”: “alex.s.gardner@jpl.nasa.gov”, “orcid”: “0000-0002-8394-8889”}
Author3 = {“name”: “Mark Fahnestock”, “affiliation”: “University of Alaska Fairbanks”, “email”: “mfahnestock@alaska.edu”, “orcid”: “”}
Author4 = {“name”: “Ted Scambos”, “affiliation”: “University of Colorado Earth Science and Observation Center”, “email”: “tascambos@colorado.edu”, “orcid”: “”}
Author5 = {“name”: “Maria Liukis”, “affiliation”: “NASA Jet Propulsion Laboratory”, “email”: “maria.liukis@jpl.nasa.gov”, “orcid”: “”}
Author6 = {“name”: “Chad Greene”, “affiliation”: “NASA Jet Propulsion Laboratory”, “email”: “chad.a.greene@jpl.nasa.gov”, “orcid”: “”}
Author7 = {“name”: “Yang Lei”, “affiliation”: “California Institute of Technology”, “email”: “ylei@caltech.edu”, “orcid”: “”}
Author8 = {“name”: “Joe Kennedy”, “affiliation”: “University of Alaska ASF”, “email”: “jhkennedy@alaska.edu”, “orcid”: “”}
Author9 = {“name”: “Bruce Wallin”, “affiliation”: “National Snow and Ice Data Center”, “email”: “bwallin@nsidc.org”, “orcid”: “0000-0002-4928-1814”}
Table of Contents¶
1.2 Purpose ¶
The itslive-explorer notebook allows users to explore and visualize worldwide glacier surface velocity using data produced by NASA’s JPL autonomous Repeat Image Feature Tracking algorithm (Gardner et al., 2018).
Because of its high temporal and spatial resolution, ITS_LIVE data amounts to 8+ million NetCDF files (and growing) stored in AWS S3. For the users’ convenience, exploration and filtering of this data can be done using an ipyleaflet-based widget that leverages the ITS_LIVE search API. After relevant data granules for an area are downloaded (for example a glacier of interest), the notebook provides users with an xarray-powered method to build a data cube ready for time series analysis and from which valuable scientific insights can be gathered.
1.3 Technical contributions ¶
Demonstration of time series analysis using the ITS_LIVE velocity pairs dataset
The main contribution of the notebook is to provide users with a simple, yet powerful, way to access and process glacier surface velocity data. The notebook uses the ITS_LIVE search API to retrieve a list of granules of interest stored on AWS S3. This can be done using the client library or using an ipyleaflet-based widget. The core example of this notebook creates a time series datacube that enables scientists to worry less about data wrangling and more about the science.
Development of underlying search API that is exposed in the notebook
The ITS_LIVE search API hosted at NSIDC is a FastAPI application that ingests geojson metadata produced by the autoRIFT processing pipeline, indexes them using PostGIS, and exposes an efficient spatio-temporal searching API.
Contributing back to the open source community
Since a considerable number of glaciers are in polar latitudes, visualization of such glacier boundaries in the common equatorial-centered map projections often lead to distortions and visual overlap/crowding. For this reason we implemented custom map projections for the ipyleaflet map widget. Also, ITS_LIVE wanted to give users who are not familiar with APIs and Python a way to get the information to their machine and begin analyzing, regardless of whether they wish to work locally from a laptop or in the cloud. Using this notebook, a user can search, download, and visualize data without the need to code a single line of Python, or on the other hand, its code and API can be used as a jumping off for developing new or existing workflows for scientists.
1.4 Methodology ¶
Data Processing¶
ITS_LIVE aims to use a cloud native approach to generate and analyze data. Leveraging the fact that the ITS_LIVE datasets are stored in AWS S3, all the processing occurs on AWS infrastructure using the hyp3 pipeline developed by the Alaska Satellite Facility. A dockerized implementation of autoRIFT is applied to input files in parallel, generating output files which are stored back in S3 along with geojson metadata for each granule.
ITS_LIVE’s autoRIFT algorithm can be applied to optical and radar imagery to detect and measure horizontal displacements between features, for example, between two repeat satellite images as a result of glacier flow, sea ice drift, large earthquake displacements, and land slides. Currently we use Landsat optical and Sentinel optical and radar imagery as input sources for glacier velocity extraction.
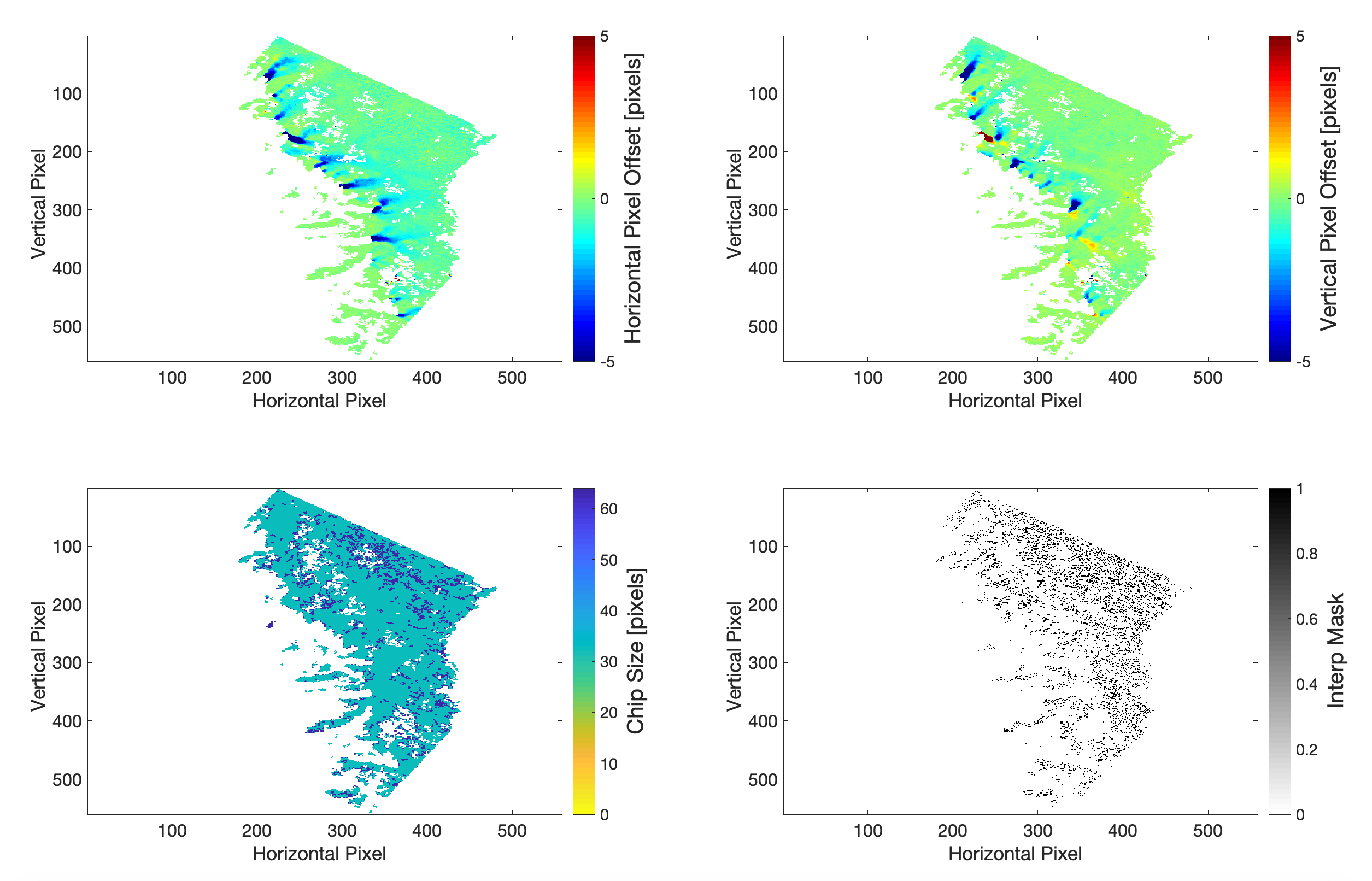
Image pairs collected from the same satellite position (“same-path-row”) are processed if they have a time separation of fewer than 546 days. This approach was used for all satellites in the Landsat series (L4 to L8). To increase data density prior to the launch of Landsat 8, images acquired from differing satellite positions, generally in adjacent or near-adjacent orbit swaths (“cross-path-row”), are also processed if they have a time separation between 10 and 96 days and an acquisition date prior to 14 June 2013 (beginning of regular Landsat 8 data). Feature tracking of cross-path-row image pairs produces velocity fields with a lower signal-to-noise ratio due to residual parallax from imperfect terrain correction. Same-path-row and cross-path-row preprocessed pairs of images are searched for matching features by finding local normalized cross-correlation (NCC) maxima at sub-pixel resolution by oversampling the correlation surface by a factor of 16 using a Gaussian kernel. A sparse grid pixel-integer NCC search (1/16 of the density of full search grid) is used to determine areas of coherent correlation between image pairs. For information on signal identification see the Normalized Displacement Coherence (NDC) filter described in Gardner et al. (2018). Information from the sparse search is then used to inform a dense search for areas with coherent correlation.
Fig 2 shows vertical and horizontal pixel displacements being correlated to create a final velocity field shown in units of meters per year.
ITS_LIVE velocity pair granules¶
The velocity granule is distributed in NetCDF format.
Coverage: All land ice
Date range: 1985-present
Resolution: 240m
Projections: UTM zones for mid latitudes, EPSG:3031 for Antarctica, and EPSG:3413 for northern latitudes.
Scene-pair separation: 6 to 546 days
Version: Beta, V0.0
Granule Search and Analysis¶
The ITS_LIVE search API has 2 endpoints to retrieve a list of granules of interest using the OpenAPI specification:
/velocities/coverage/gets an aggregate by year (a faceted result) of how many granules will be returned given the user’s spatiotemporal parameters
/velocities/urls/produces a list of S3 file URLs that match the user’s spatiotemporal parameters
1.5 Results ¶
ITS_LIVE seeks to minimize remaining barriers to using satellite observations for cryosphere research. This includes the production, synthesis, delivery and analysis of such data. The project follows an open-source philosophy with open data and code. In the 2 and a half years since its inception, ITS_LIVE has already supported thousands of unique users, provided nearly 10TB of science data and contributed to more than 50 publications.
ITS_LIVE’s cloud native approach used to create our dataset makes it ready to be scaled to handle the tsunami of remote sensing data NASA and other agencies are projecting to gather in the coming few years. This notebook is presented as a central collaborative access point to get the data and start working on the science right away.
1.6 Funding ¶
Award1 = {“agency”: “NASA”, “award_code”: “Making Earth System Data Records for Use in Research Environments (MEaSUREs) Program”, “award_URL”: “https://earthdata.nasa.gov/esds/competitive-programs/measures/its-live”}
1.7 Keywords ¶
keywords=[“glacier”, “surface”, “velocity”, “dataset”, “nasa”]
1.8 Citation ¶
Gardner, A. S., M. A. Fahnestock, and T. A. Scambos, 2021. ITS_LIVE Regional Glacier and Ice Sheet Surface Velocities. Data archived at National Snow and Ice Data Center; doi:10.5067/6II6VW8LLWJ7.
1.9 Work In Progress - improvements ¶
The current analysis workflow still relies in a dated “download and analyze” paradigm but incorporates ideas and functionality made practical with Pangeo, such as processing the granules on a Dask cluster. A dedicated Dask cluster for our users is out of scope for ITS_LIVE, however it would be straightforward to adapt the current client library and examples to use one.
To-Do:¶
1. On demand cube generation(back-end): An intermediate step between the current workflow to analyze the velocity granules locally and in the cloud is to implement the data cube generation on the ITS_LIVE back-end. This way the users will only care about downloading the slice of data they need with the variables they need. This sliced cube could also be generated by Harmony at some point, see point no. 4.
2. Velocity basemap: the current map widget does not have a basemap that reflects the global velocity mosaics. The trick is to adapt gdal2tiles or something like it to process enough regional maps into a partition that is compatible with NASA GIBS grid definitions(the basemaps used in the widget)
3. Include elevation change datasets: Velocity is not the only ingredient scientist need in order to analyze glacier movements, elevation change data is also part of ITS_LIVE but is not yet included on the current notebook. Once we have both datasets, more accurate and interesting analyses will be possible e.g. mass balance trends.
4. Harmony integration: NASA’s Harmony is NASA’s next generation data processing tool that will allow scientists to get subsetted and analysis ready data in cloud data format. Having ITS_LIVE as a Harmony compatible data will close the cloud native circle stated earlier.
1.10 Acknowledgements ¶
This effort was funded by the NASA MEaSUREs program in contribution to the Inter-mission Time Series of Land Ice Velocity and Elevation (ITS_LIVE) project (https://its-live.jpl.nasa.gov/) and through Alex Gardner’s participation in the NASA NISAR Science Team.
NSIDC allocated time to the project as well as computing resources via the DAAC agreement.
Datasets used
Landsat 4,5,7,8 data were provided by the U.S. Geological Survey.
Copernicus Sentinel-1 and Sentinel-2 data were acquired, processed, and generated by the European Space Agency (ESA).
ERS-1 and ERS-2 altimetry data were provided by ESA’s Reprocessed ESA ERS Altimetry (REAPER) project.
Envisat and CryoSat altimetry data were acquired, processed, and generated by the European Space Agency (ESA).
ICESat altimetry data was provided by NASA through NSIDC.
2. Setup ¶
There are 2 ways of using this notebook, we are going to use code but after all the cells are executed, we would like the users to try the same approach with the widget.
2.1 Library import ¶
# Code cell No. 1
# data manipulation and plotting.
import xarray as xr
# ITS_LIVE search client which can be used as a widget or just code.
from SearchWidget import map
from VelocityProcessing import VelocityProcessing as vp
# horizontal=render in notebook. vertical = render in sidecar
m = map(orientation='horizontal')
3. Parameter definitions ¶
ITS_LIVE data come from multiple scenes and satellites, which means a lot of overlap. In this case all the scenes that match with our spatial criteria will be returned. Fig 4 shows overlaps of different Landsat scenes on a given zone.
Note: This notebook was designed to build data cubes on a glacier scale rather than larger areas. we will use Pine Island Glacier for demo purposes.
3.1 Search parameters ¶
polygon/bbox: LON-LAT pairs separated by a comma.
start: YYYY-mm-dd start date
end: YYYY-mm-dd end date
min_separation: minimum days of separation between scenes
max_separation: maximum days of separation between scenes
percent_valid_pixels: % of valid pixel coverage on glaciers
serialization: json,html,text
You can use Python or CURL to get the granules list. The OpenAPI endpoint can also be used to access the data.
https://nsidc.org/apps/itslive-search/docs
or with Curl
curl -X GET "https://nsidc.org/apps/itslive-search/velocities/urls/?polygon=-68.0712890625%2C-69.77135571628376%2C-65.19287109375%2C-69.77135571628376%2C-65.19287109375%2C-68.19605229820671%2C-68.0712890625%2C-68.19605229820671%2C-68.0712890625%2C-69.77135571628376&start=2000-01-01&end=2020-01-01&percent_valid_pixels=80" -H "accept: application/json"
# Code cell No. 2
# Pine Island Glacier
params = {
'polygon': '-101.1555,-74.7387,-99.1172,-75.0879,-99.8797,-75.46,-102.425,-74.925,-101.1555,-74.7387',
'start': '1984-01-01',
'end': '2020-01-01',
'percent_valid_pixels': 10,
'min_separation': 6,
'max_separation': 180
}
# Search populates the "filtered_urls" variable in our class instance, next we can actually filter this list to limit the number of granules we download.
granule_urls = m.Search(params)
print(f'Total granules found: {len(granule_urls)}')
3.2 Tunning our search ¶
Besides the spatio-temporal parameters, the other important parameter to take into account is percent_valid_pixels. Old scenes form the 80s don’t have a lot of good quality coverage and lowering this parameter may return more usable results. For recent years, however, quality is better and lowering it may return too many results.
# Code cell No. 3
# Pine Island Glacier with a higher minimum coverage treshold and max separation to 120 days
params = {
'polygon': '-101.1555,-74.7387,-99.1172,-75.0879,-99.8797,-75.46,-102.425,-74.925,-101.1555,-74.7387',
'start': '1984-01-01',
'end': '2020-01-01',
'percent_valid_pixels': 50,
'min_separation': 6,
'max_separation': 120
}
granule_urls = m.Search(params)
print(f'Total granules found: {len(granule_urls)}')
4. Data import ¶
4.1 Data Filtering ¶
Depending on your application, more than a thousand granules may be ok, but if, for example, you only want to get an overall glance at the behavior of a particular glacier over the years, or limited to e.g. a certain time of year, you can limit the number of granules per year and/or with middates from a set of given months. With processing done either on the back-end (development in progress), or using a Dask cluster, limiting granules becomes less necessary.
# Code cell No. 4
# filter_urls requires a list of urls, the result is stored in the m.filtered_urls attribute
filtered_granules_by_year = m.filter_urls(granule_urls,
max_files_per_year=10,
months=['November', 'December', 'January'],
by_year=True)
# We print the counts per year
for k in filtered_granules_by_year:
print(k, len(filtered_granules_by_year[k]))
print(f'Total granules after filtering: {len(m.filtered_urls)}')
4.2 Downloading data ¶
We have 2 options to download data: we can download filtered urls (by year or as a whole) or we can download a whole set of URLs returned in our original search.
Single year example:
files = m.download_velocity_granules(urls=filtered_granules_by_year['2006'],
path_prefix='data/pine-glacier-2006',
params=params)
The path_prefix is the directory on which the netcdf files will be downloaded to and params is to keep track of which parameters were used to download a particular set of files.
We can also download the whole series
files = m.download_velocity_granules(urls=m.filtered_urls,
path_prefix='data/pine-glacier-1996-2019',
filtered_urls=params)
# Code cell No. 5
filtered_urls = m.filtered_urls # or filtered_granules_by_year
project_folder = 'data/pine-1996-2019'
# if we are using our parameters (not the widget) we assign our own dict i.e. params=my_params
files = m.download_velocity_granules(urls=filtered_urls,
path_prefix=project_folder,
params=params)
5. Data processing and analysis ¶
The most common use case for the velocity granules is to generate a time series. In order to create one, we need to concatenate multiple granules from the region of interest for the time we want information from. ITS_LIVE provides a processing module that will load all the velocity granules on a given directory and use a provided geojson geometry to clip the files and generate a datacube for the area.
# Code cell No. 6
# First let's open a single data granule (included in this notebook)
velocity_granule = xr.open_dataset('data/LE07_L1GT_001113_20121118_20161127_01_T2_X_LE07_L1GT_232113_20121104_20161127_01_T2_G0240V01_P059.nc')
velocity_granule
# Code cell no. 7
# xarray has built-in methods to plot our variables, in this case the velocity which is the important one in our granules.
velocity_granule.v.plot(x='x', y='y')
5.1 Building a data cube ¶
The velocity granules we downloaded overlap with Pine Island Glacier, however they also contain data outside of its boundary. This means that we will have to process these NetCDF files by first carving out our region of interest and reprojecting our of intersection into a common projection (from e.g. multiple UTM zones). These operations will allow us to stack multiple scenes and build our time series analysis. This notebook uses rio-xarray to clip and reproject data via the load_cube method.
Notes:
1.The following code cells are intended to work after multiple velocity granules from different years and the same region are downloaded.
2.
load_cubeis not a lazy function. It will allocate granules on memory in order to create the cube. This means that if we try to load 100,000 granules the kernel will most likely run out of memory.
The signature of the load_cube method is load_cube(project_folder, clip_geom=geometry, include_all_projections=False)
The parameters are:
project_folder: The file path pattern for NetCDF velocity files
clip_geom: gejson geometry dictionary, this geometry will be used to clip the files
include_all_projections: True or False, if True the cube will include granules on different UTM zones than the most common one for the selected area.
If we don’t use the map widget we can also use a handy function to get the geojson polygon from a bounding box
# code cell No. 8
# If we have a list of coordinates or a bbox we can get the correspondant geojson by
# geometry = vp.box_to_geojson([-49.79, 69.06, -48.55, 69.25])
# geometry = vp.polygon_to_geojson([(-48.55, 69.06),(-48.55, 69.25),(-49.79, 69.25),(-49.79, 69.06),(-48.55, 69.06)])
# The load_cube method needs a GeoJSON geometry to clip the region of interest.
geometry = {'type': 'Polygon',
'coordinates': [[[-101.155511, -74.738709],
[-99.117207, -75.087959],
[-99.879774, -75.46007],
[-102.42516, -74.925034],
[-101.155511, -74.738709]]]}
# if we are using the widget
# geometry = m.get_current_selection()['geometry']
geometry
%%time
# File pattern for our current search folder.
project_folder = 'data/pine-1996-2019/*.nc'
cube = vp.load_cube(project_folder,
clip_geom=geometry,
include_all_projections=True)
cube
5.2 Plotting a time series with xarray ¶
With our xarray data cube with time dimension we can easily use xarray plot capabilities to get a visualization of different time intervals, and start using averages, means, media or a custom computations. First, lets inspect histograms to discard potential outlier values.
# Histogram of velocities, useful to discard outliers.
cube.v.plot.hist()
# Now we are going to get annual means for glacier surface velocity, ta dah!
cube_yearly = cube.groupby('time.year').mean()
plot = cube_yearly.v.plot(x='x',
y='y',
col='year',
col_wrap=4,
cbar_kwargs=dict(
orientation= 'horizontal',
shrink=0.8,
anchor=(0.5, -0.8),
label='Velocity m/Year')
)
plot.fig.subplots_adjust(top=0.92)
plot.fig.suptitle("Pine Island Glacier Yearly Mean Velocity m/y",
fontsize=20, fontdict={"weight": "bold"})
Annex: ITS_LIVE search widget¶
Now that we downloaded and filtered data using the Search API, another approach we can try is to use the widget to interactively perform the search and have a collection granules downloaded over an area of interest to begin analyzing. Similarly, another idea is to extend this widget into an end to end tool using e.g. ipyleaflet-xarray to plot slices of the time series.
# This will display the map widget
m.display()
6. References ¶
autoRIFT: A highly accurate and efficient algorithm for finding the pixel displacement between two radar or optical images
NSIDC notebook: A Jupyter notebook to search and download ITS_LIVE scene-pair velocity.
Chad Greene’s Matlab collection: A set of Matlab functions for accessing, analyzing, and plotting ITS_LIVE velocity data. These functions are intended to streamline the process of loading ITS_LIVE mosaics, interpolating, generating flowlines, and creating maps of ice flow.
ITS_LIVE API: An API for searching ITS_LIVE scene-pair velocities.
Geogrid: A Python module for precise mapping between (pixel index, pixel displacement) in imaging coordinates and (geolocation, motion velocity) in geographic Cartesian (northing/easting) coordinates
Pangeo: A community platform for Big Data geoscience